 KONECT ‣ Networks ‣
Buy Me a Coffee
KONECT ‣ Networks ‣
Buy Me a Coffee
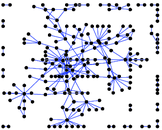
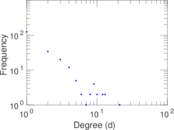
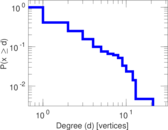
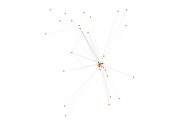
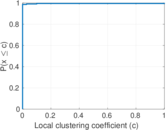
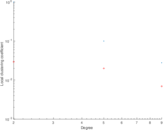
This is a network of protein–protein interactions from PDZBase.
| Code | MP
| |
| Internal name | maayan-pdzbase
| |
| Name | PDZBase | |
| Data source | http://research.mssm.edu/maayan/datasets/qualitative_networks.shtml | |
| Availability | Dataset is available for download | |
| Consistency check | Dataset passed all tests | |
| Category | Metabolic network | |
| Node meaning | Protein | |
| Edge meaning | Interaction | |
| Network format | Unipartite, undirected | |
| Edge type | Unweighted, no multiple edges | |
| Loops | Contains loops |
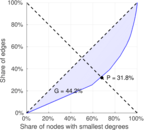
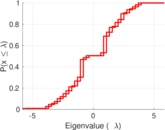
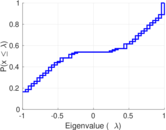
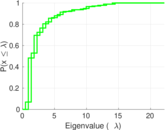


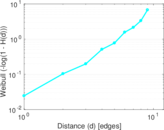
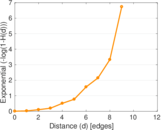


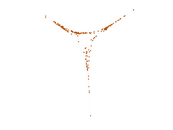
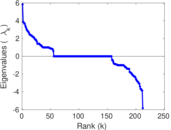
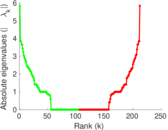
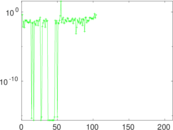
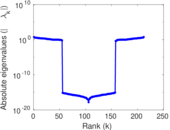
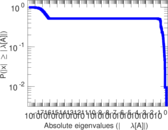
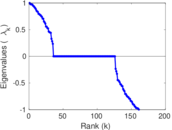
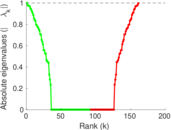

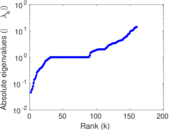
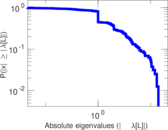
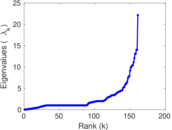
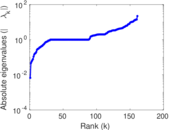
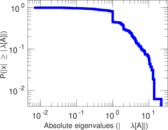
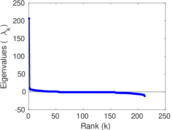
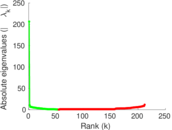
| [1] | Jérôme Kunegis. KONECT – The Koblenz Network Collection. In Proc. Int. Conf. on World Wide Web Companion, pages 1343–1350, 2013. [ http ] |
| [2] | Thijs Beuming, Lucy Skrabanek, Masha Y. Niv, Piali Mukherjee, and Harel Weinstein. PDZBase: A protein–protein interaction database for PDZ-domains. Bioinformatics, 21(6):827–828, 2005. |